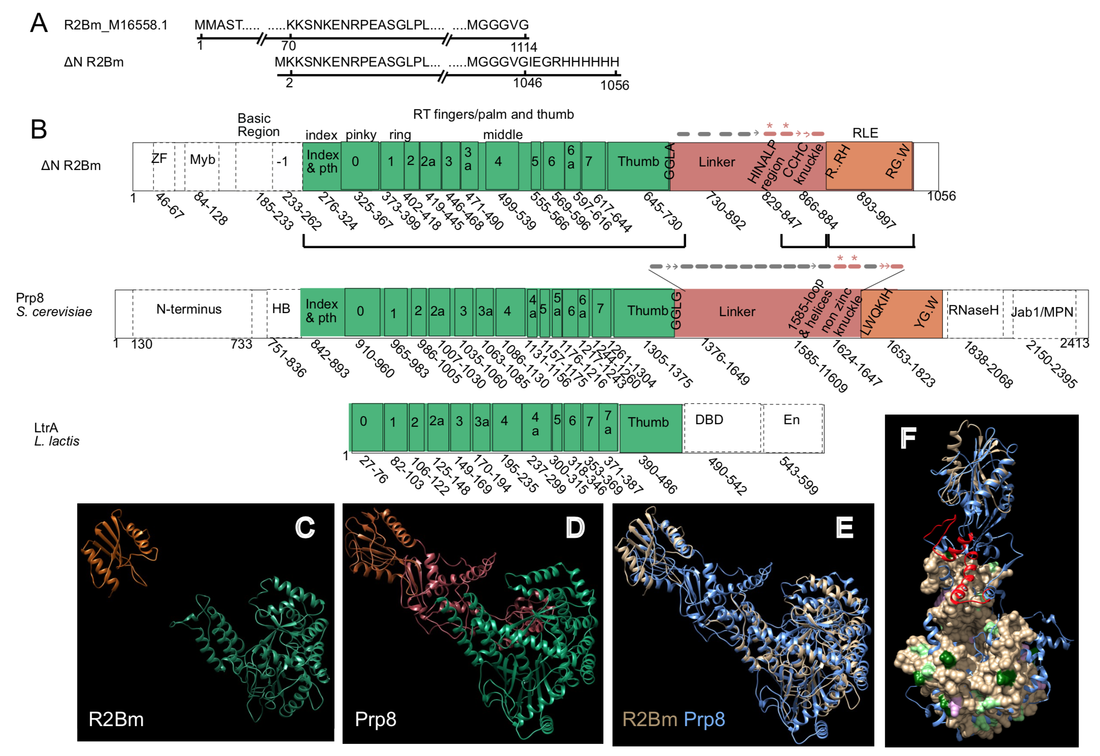
ABOUT THE CHRISTENSEN LABNon-LTR RetrotransposonsRetrotransposable elements are selfish DNA elements that replicate within a host genome through an RNA intermediate. Retrotransposable elements have been found residing in the genomes of most eukaryotic organisms studied to date, often accounting for a sizable fraction of the host genome and are responsible for most, if not all, of the reverse transcribed DNA in the genome. Long interspersed elements (LINEs), also called non-LTR retrotransposons, are a major class of retrotransposable elements (Figure 1). LINEs are generally inherited from parent to offspring (vertical inheritance) and replicate during the formation of germ line cells. LINE-induced chromosomal aberrations can result in genetic diseases in the resulting offspring and can generate new host genes or regulatory sequences over evolutionary time scales. LINEs are also known to mobilize in various somatic cells, such as in neuronal development and during the progression of many cancers. The machinery of LINEs is used by short interspersed elements (SINEs), parasites of LINEs, to replicate and is responsible for pseudogene formation. It is important from medical and evolutionary perspectives to understand how LINEs insert into the host genome (Figure 2).
|
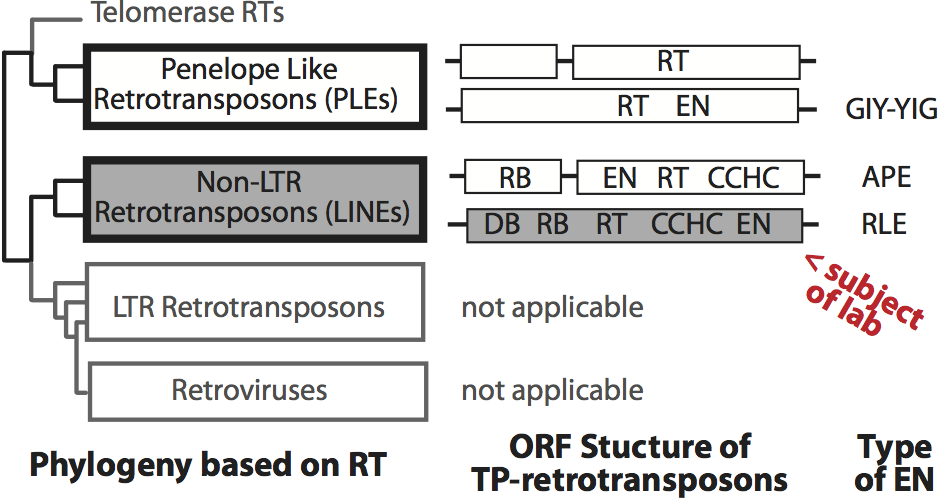
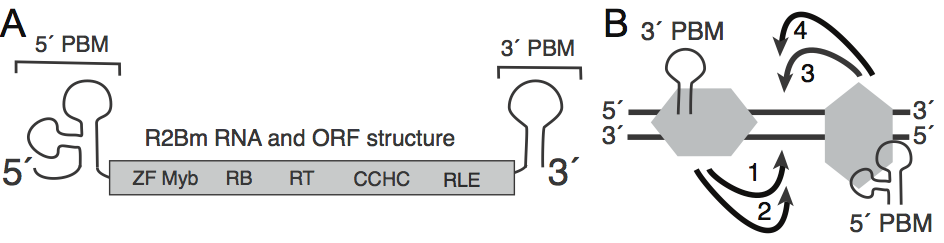
Figure 1: Phylogeny of reverse transcriptase bearing elements and the open reading frame (ORF) structure of target primed retrotransposons.Figure 2: Structure of the R2 element and the TPRT mechanism. Panel A is the RNA and ORF structure of the R2 element from Bombyx mori. RNA motifs present in the 5′ and 3′ untranslated regions bind R2 protein and are marked as 5′ and 3′ protein binding motifs (PBMs), respectively. Panel B is the four-step integration model: (1) DNA cleavage of the bottom strand, (2) TPRT, (3) DNA cleavage of the top strand and (4) second strand DNA synthesis.
|
Recent AdvancesWe have had an exciting couple of years. Big advances have been made with even more in the pipeline!
|